This document contains developer documentation for BioImage Suite Web. Follow this link for the end-user documentation/manual.
A web page version (as opposed to browsing the source) of this page can be found at this link.
Introduction
BioImage Suite Web (BisWeb) is a web-based medical image analysis tool geared towards processing neural images. We gratefully acknowledge support from the NIH Brain Initiative under grant R24 MH114805 (Papademetris X. and Scheinost D. PIs). A good overview of the software can be found in slides from a presentation at the 2018 NIH Brain Initiative Meeting, which was the first public introduction of the software.
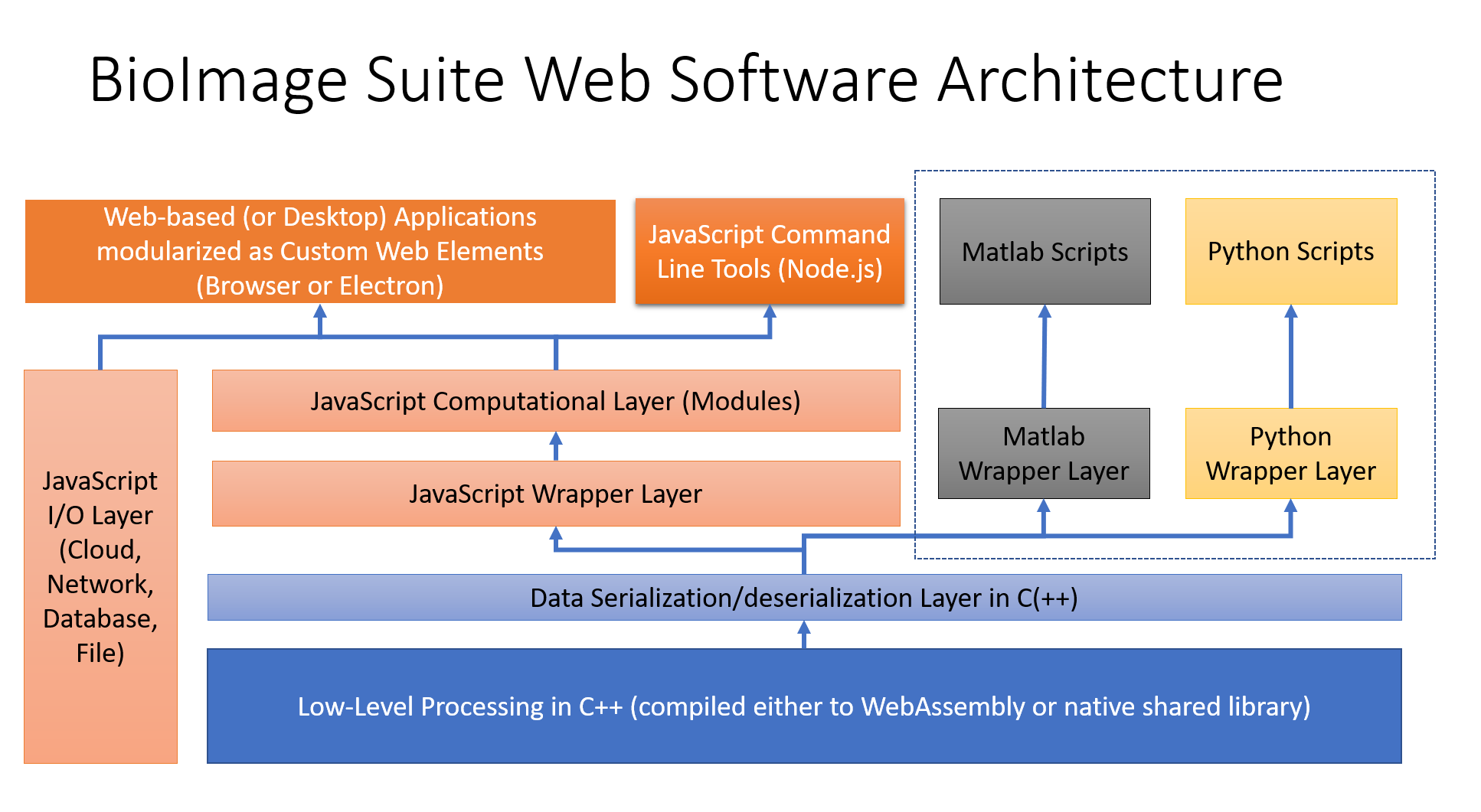
The architecture of BioImage Suite Web is shown below:
BioImage Suite Web uses a mixture of JavaScript for the user interfaces and C++ for the image processing with optional Python and Matlab wrappers. Additional information about aspects of the software can be found in other files in this directory, such as:
- Some background material on why JavaScript is the primary language for Biomage Suite Web in WhyJS.md.
- Aspects of JavaScript that are particularly relevant to the project in AspectsofJS.md.
- How to program using the base BioImage Suite Web Libraries in JS in BisWebJS.md.
- The interface from JavaScript to WebAssembly in JStoWASM.md.
- Using the base BioImage Suite Web libraries from Python in BisWebPython.md.
- Using the base BioImage Suite Web libraries from Matlab inBisWebMatlab.md. This code is primitive compared to the rest of the code base and more importantly is unsupported. Use at your own risk!
- Descriptions of the module architecture in two separate documents
- JavaScript: ModulesInJS.md
- Python: ModulesInPython.md.
- Details about how the Electron desktop applications are constructed in DesktopAppsWithElectron.md.
Setting up your Development Environment
The goal of this section is to help the reader set up a proper software development environment. BioImage Suite Web uses the following tools:
- Git : A popular version control system.
- Node.js : The command line JavaScript interpreter. Packaged with npm which is the package manager that comes with node.js.
- Gulp: An automation tool designed to ease JavaScript build tasks.
- webpack: A packaging tool that enables Node.js style modules in browser applications.
- Mocha: A scripting library designed to automate regression testing.
- JSHint: “A tool that helps to detect errors and potential problems in your JavaScript code” (from the webpage).
- JSDoc: A parsing tool that generates documentation from comments in code.
- Electron: A tool that uses the HTML/CSS/JS that makes up a webpage to create a desktop application.
Editing code also requires a text editor. Users with no prior experience may want to try Microsoft’s Visual Code editor. Sublime and Atom are also fine choices. Emacs or Vim may be considered for the truly dedicated/crazy, but users of such esoterica tend to know who they are already.
More information about each of these tools may be found in the links contained in this section.
Setting up your Development Environment
Option 1. Docker Environment (Advanced)
If you are comfortable with Docker, then you can create and build bisweb in a docker environment. See our docker repository.
In particular you will need to
-
Install the container
docker pull xeniosp/bisweb
-
Log in to the container
sudo docker run -p 8080:8080 -it xeniosp/bisweb bash
-
The source tree for bisweb will be in the directory /root/bisweb/src and /root/bisweb/gpl (for the gpl plugin). See the Dockerfile for more information. You can update the source (this is mapped to
develbranch) usinggit pullas usual.
Option 2. Linux/Ubuntu
Compiling BisWeb requires a number of prerequisites.
For Ubuntu you will need to run the following commands (unless these packages are already on your system)
sudo apt-get -yqq update
sudo apt-get install -yqq python-pip python-dev python3 python3-pip unzip g++ gcc cmake cmake-curses-gui
sudo apt-get install -yqq doxygen graphviz
sudo apt-get install -yqq curl openjdk-8-jdk git make dos2unix
sudo curl -sL https://deb.nodesource.com/setup_10.x | sudo bash
sudo apt-get install -yq nodejs
Then install the following 2 python packages (if you are interested in python)
sudo pip3 install numpy nibabel
Then install the following npm dependencies:
sudo npm install -g gulp mocha rimraf
sudo npm install -g electron --unsafe-perm=true --allow-root
sudo npm install -g electron-packager
These steps are identical to what is used for the Docker-based devel setup described above.
Aside: Microsoft Windows
We suggest using the Windows Subsystem for Linux (WSL). Install the Ubuntu VM and follow the same steps as for Linux/Ubuntu above.
Option 3. MacOS 10.14
This is similar to Linux. First install Xcode and homebrew if you do not already have these installed.
Then install the pre-requisite packages using brew – the instructions below are for Node V.10.
brew install python2 python3 nodejs cmake node@10 doxygen graphviz
brew link python2
brew link --force node@10
brew cask install java
Then install the python and npm dependencies as in the Ubuntu case using:
pip3 install numpy nibabel
npm install -g gulp mocha rimraf
npm install -g electron --unsafe-perm=true --allow-root
npm install -g electron-packager
Building and Running BioImage Suite Web
BioImage Suite Web is currently compiled/packaged in directories inside the source tree. The contents are as follows:
- node_modules — The location of dependent node_modules. This will be populated by npm.
- build — The core build directory.
- build/wasm — The build directory for the WebAssembly code. You can move this out if your prefer; however, the key output files will be redirected to the build directory so that webpack can find them.
- build/dist — The output directory for
.zipfiles or packaged Electron applications. - build/native — (optional) This is where the native libraries for accessing the C++ code from Python and Matlab will be built.
- build/web — The output directory for creating the final web-applications and associated
.css,.htmlfiles etc.
- compiletools — Scripts and configuration files to help with compiling.
- config — Configuration files for webpack, JSDoc etc.
- cpp — C++ code.
- doc — Directory containing this and other documentation files.
- docker — The docker configuration files
- js — Directory containing all js code
- lib — Directory containing js external libraries and some css/html that are either not distributed via npm or are customized in some way.
- matlab — Directory containing the primitive Matlab wrapper code.
- python — Directory containing the Python code.
- test — Directory containing the regression tests and data.
- various — Miscellaneous files.
- various/download — Versions of Emscripten (this is a skeleton version) and Eigen for installation.
- various/wasm — Pre-built version of the Wasm-related JS files to expedite getting started. These can be copied to
build/wasmto eliminate the need to compile C++ to Web Assembly. These are updated periodically though so to use up-to-date code they will have to be recompiled. - various/config — Configuration files for Emacs, Bash and Visual Studio Code that may be useful.
- web — The directory containing the
.html/.cssfiles for the web and desktop applications. This also contains the configuration files for Electron desktop applications.
In addition there are two key files that live in the main directory
package.json— The package description file for npm that lists the dependencies of our software. Generates a companion file,package-lock.jsongulpfile.js— The configuration file for Gulp.
Getting the BioImage Suite Web code
First create an empty director. Avoid paths that have spaces in them; a folder named ‘bisweb’ or something similar in the home directory will work well.
cd ~
mkdir bisweb
git clone https://github.com/bioimagesuiteweb/bisweb bisweb
You should also get the gpl plugin
git clone https://github.com/bioimagesuiteweb/gplcppcode gpl
The source code for the project may be found on Github.
Building the Code
Assuming the steps above, the BioImageSuite Web code should be inside a folder
named bisweb. This will be used as the name of the root directory for the
project, but you can name it whatever you’d like so long as you’re consistent.
cd bisweb
Note the presence of package.json. This contains references to the dependencies required by BioImageSuite Web. Ensure that these are up to date by typing:
npm install
or, for more verbose output:
npm install -d
Note: Dependencies may change over time. If Bisweb does not perform as expected try checking if there are updates to the dependencies.
Note 2: If npm install fails to install tensorfow.js (probably because you
do not have a proper node-gyp) setup, simply delete the line containing
tensorflow from package.json and try again. This is optional at this point.
To create the WebAssembly binaries and build folder structure from the
source files, use the createbuild.js script
node ./config/createbuild.js
This will create a number of sub-directories, (e.g. build/web, build/wasm,
build/dist, build/native, build/doc build/install) and also install
emscripten as needed.
Then you can perform a full initial build using
cd build
./fullbuild.sh
Open the Web Applications
To do this type:
gulp
This does three things:
- It creates a light-weight web server.
- It runs JSHint that performs syntax checking on all the JS files
- It runs webpack which packages all the
.jsfiles into a single web-compatible JS output
Open a web-browser and go to http://localhost:8080/web. If it all works the main screen should pop up.
Note: The observant reader may notice that the .html files live in the web directory. Executing gulp build modifies these and places them in build/web for eventual distribution/packaging. This does some re-writing of the HTML header to change paths etc.
Running Regression Tests
The regression tests should function by this point. These should be executed to test proper integration of the WebAssembly code. To do so, type:
cd bisweb/test
mocha test
This will take a few minutes to finish. At this point the JS-development directory should be fully functional.
Under the Hood
The fullbuild.sh script calls 4 container scripts
wasmbuild.sh– this builds the C++ code as a WebAssembly Librarywebbuild.sh– this builds the JS bunles for the Web application (this depends on wasmbuild.sh)nativebuild.sh– this builds the Python and Matlab bindings (this also depends on wasmbuild.sh being run at least once)testbuild.sh– this runs a small subset of the regression tests.
If you simply want to rebuild one of these components, just run the individual
script (e.g. webbuild.sh to rebuild the web application)
Configuring and Building – The Manual Way
The WASM Code
This requires some understanding of CMake. There is lots of info online on this.
You will need to run cmake to configure the project as a “cross-compiled” application. The easiest way to do this (if you follow our instructions completely) is as follows:
cd build/wasm
Then to accept all defaults type
../cmake.sh .
Note: If you would like to customize things replace cmake.sh with ccmake.sh. to use the ccame GUI version of cmake.
The core settings are
- BIS_A_EMSCRIPTED – if ON cross compile using Emscripten else build native library using native compilers (this is how the Python libraries are compiled)
- BIS_WEB_OPT – this is the optimization flags for the compiler (-O2 is default)
- BUILDNAME – this is the automatically generated build name
- BUILD_TESTING – set this to ON to enable ctest-style testing (in addition to mocha). CTest calls mocha.
- EXECUTABLE_OUTPUT_PATH – Make sure this goes to
build/wasmas this is wherewebpackand thenode.jswill look for the final libraries. - EIGEN3_DIR – set this to the location of the eigen cmake directory – it should be set automatically for you.
This cmake.sh/ccmake.sh scripts simply sets some environment variables and runs cmake with the necessary flags. You can can do this manually and set the flags inside cmake but this is not for the faint-hearted.
One CMake is done, on the console simply type
make
or better use the -j flag to set multiple parallel jobs
make -j2
If it all goes well you will have a fresh set of files in your build/wasm directory:
- libbiswasm_wrapper.js – the JS wrapper code (more on this in another document)
- libbiswasm.wasm – the actual web assembly output file (which we do not use directly)
- libbiswasm.js – the Emscripten Module that packages the web assembly library
- libbiswasm_wasm.js – a custom script that embeds the WASM file.
Note: In the future, if you make any changes to the C++ code (or inherit some via git pull), source setpaths.sh before typing make as this may invoke cmake which will not find (by default) the paths to Emscripten.
In addition you will have created the command line tools in build/wasm/lib
This is a set of bash and batch files plus two large-ish js files bisweb.js and bisweb-test.js that are the webpack-packaged versions of the command line code. You can install and package these in the usual cmake way using make install and make package if you know what to set up the proper configuration in CMake.
If you are feeling brave you can at this point type:
make test
This will run the regression tests. It will take a while but hopefully everything passes.
Building the C++ Code as a Native Shared Library for Python/Matlab
You will need to install Python 3.5 or higher for this to work.
This is simpler than the WebAssembly build BUT you need to have the WebAssembly libraries compiled first as the Python module descriptions (more on this in ModulesInPython.md) are created from the JS Code.
If you followed the steps above you will have a directory called build/native. Then cd to this and run the script cmake_native.sh as follows:
cd build/native
../cmake_native .
(Again if you would like to customize things replace cmake_native.sh with ccmake_native.sh.)
Once you are done type:
make -j2
and then
make test
as before.
Creating the Code Documentation
For the JS code simply type
gulp jsdoc
This will create a set of html files in build/doc.
For the C++ code you will need to install doxygen and dot.l On Ubuntu this can be done as:
sudo apt-get install doxygen graphviz
On Mac OS (using brew) you can similarly get these using:
brew install doxygen graphviz
Then simply type
gulp cdoc
The final output will go in the directory build/doc/doxygen/html.
Installing and Packaging
Web Applications
The first is for the webpage itself. To do this type
gulp build -m
gulp zip
The setting -m turns on minification of the JS code and turns off debug statements. The final result should be a zip file in build/dist that can simply be uploaded to a web server.
Electron Application
To package bisweb as a desktop application for electron simply type
gulp package
You will find the resulting file (.app,.exe,.zip depending on the platform) in build/dist.
Command Line Tools
For either Python tools or WASM commandline tools you can install these using
make install
This will send the files to the directory CMAKE_INSTALL_PREFIX/bisweb. You can set the value of CMAKE_INSTALL_PREFIX in cmake, the default is /usr/local.
You can also configure packaging using cpack within CMake. Then type:
make package
This will create the appropriate .tar.gz, .sh, .zip (depending on options selected) file for you to share.
A cool little trick is to make install both the WASM and the Python tools to the same directory. Then simply zip this and you will have a single JS/Python command line installation!
Dropbox and Google Drive Keys
The only source file that is not publicly supplied is the file containing keys for Google Drive and Dropbox access. A skeleton files is provided in js/nointernal/bis_keystore.js. If you intend to include this code in your own application you will need to register it with Dropbox/Google Drive as needed and obtain your own keys and add them to this file (which you should also keep private). The easiest way to do this in the current source setup is to:
- Create a directory structure that contains two folders
- bisweb (or anything you want) – clone the BioImage Suite Web repository inside this directory.
- internal (this must be called internal) – this is for your own private code.
-
In internal create a subdirectory called
js -
Copy the files from
bisweb/js/nointernaltointernal/jsdirectory and edit them to set the correct keys. -
Run the build using
cd bisweb gulp --internal 1
The parameter “–internal 1” instructs webpack to include the code from the internal directory and not include the boilerplate from js/nointernal – see config/webpack.config.js if you are curious as to how this happens.
Web-based Tests
Browser:
To run tests in the browser (this applies only to the module tests) type
gulp serve
Then navigate to:
http://localhost:8080/web/biswebtest.html
Then select the tests to run and click Run Tests to execute.
Electron:
To run tests in Electron (this applies only to the module tests) type
gulp build
Then (assuming you are in the src directory) type:
electron web biswebtest
Then select the tests to run and click Run Tests to execute.
This page is part of BioImage Suite Web. We gratefully acknowledge support from the NIH Brain Initiative under grant R24 MH114805 (Papademetris X. and Scheinost D. PIs, Dept. of Radiology and Biomedical Imaging, Yale School of Medicine.)