Image Processing Tools In BioImage Suite Web
The applications in BioImage Suite Web can do many standard medical image analysis processing tasks. These are available from the Image Processing menu of the main orthogonal viewer application and the dual viewer applications.
Tools Summary
In the Orthogonal Viewer Tool these appear as:
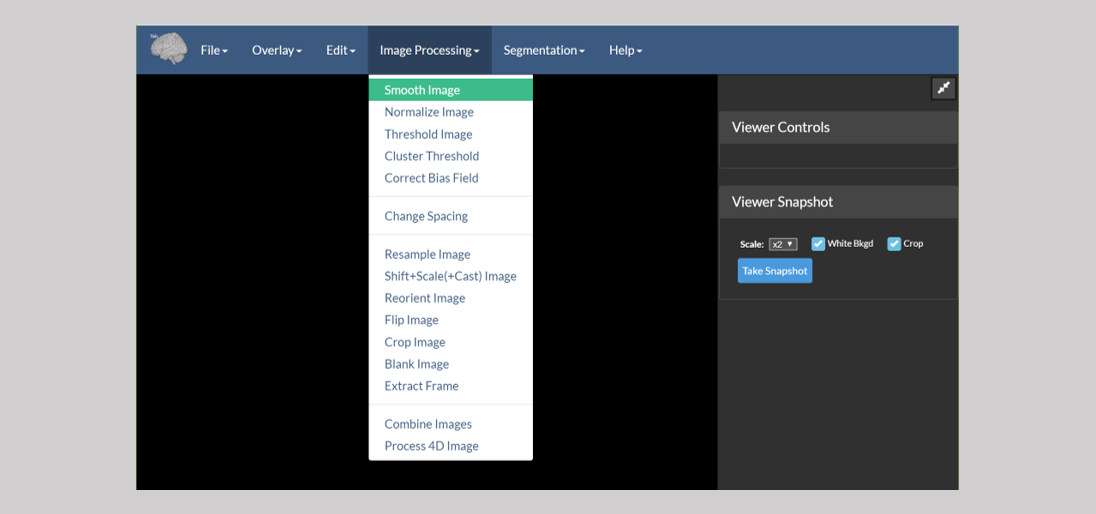
These tools perform the following tasks:
- Smooth Image — Performs a Gaussian blur with kernel size and sigma set by the user.
- Normalize Image — Normalizes the intensity range of the image. Generally used to proprocess images for registration, though note that the registration tools in BioImage Suite do this automatically.
- Threshold Image — Performs either binary or band-pass thresholding.
- Cluster Threshold Image — Thresholds to form clusters in order to elimninate small bright collections of voxels.
- Correct Bias Field — Corrects slice-inhomogenerity to alleviate B1-bias field artifacts in MRI images.
- Change Spacing — Changes the voxel spacing (essentially the resolution) in the header of an image — the image is not changed in any way, only the interpratation of the header. This is often needed when the image was saved incorrectly from another source, e.g. a faulty Matlab script.
- Resample Image — Resamples an image to a new resolution.
- Shift+Scale+(Cast Image) — Does two things: (i) manipulates the image intensities e.g. Y = scale * (X + shift) and (ii) changes the image type, e.g. to short or to float.
- Reorient Image — Reorients a medical image to a standard orientation (i.e. RAS, LPS, or LAS). The reorientation is simply performed by permuting the axes. No interpolation or resampling is done.
- Flip Image — Flips an image along any combination of the
i,jandk-axes. - Crop Image — Crops an image to size. This can be done in 4D to extract a subset of the frames in a 4D image. It can also be used to pad an image if negative bounds are used.
- Blank Image — Crops an image, but instead of changing the image’s size, the
outsideregion is simply blanked (zeroed-out). - Extract Frame — Extracts a single frame from a 4D image.
- Combine Images — Mathematically combines two images, i.e. add, substract, multiply, divide, or appends them.
- Process 4D Image — Processes a 4D image along its time axis. For example, it computes the mean frame and other similar measures.
Note: All of these modules can be run on the command line as well — see the Command Line Tools document for more information. You can also regression test the modules — see the Testing document for more on this.
Using a Tool — Single Viewer Mode
All the tools listed above can run on the viewer. Consider the case of the Flip Image tool, as explained in the figure below.
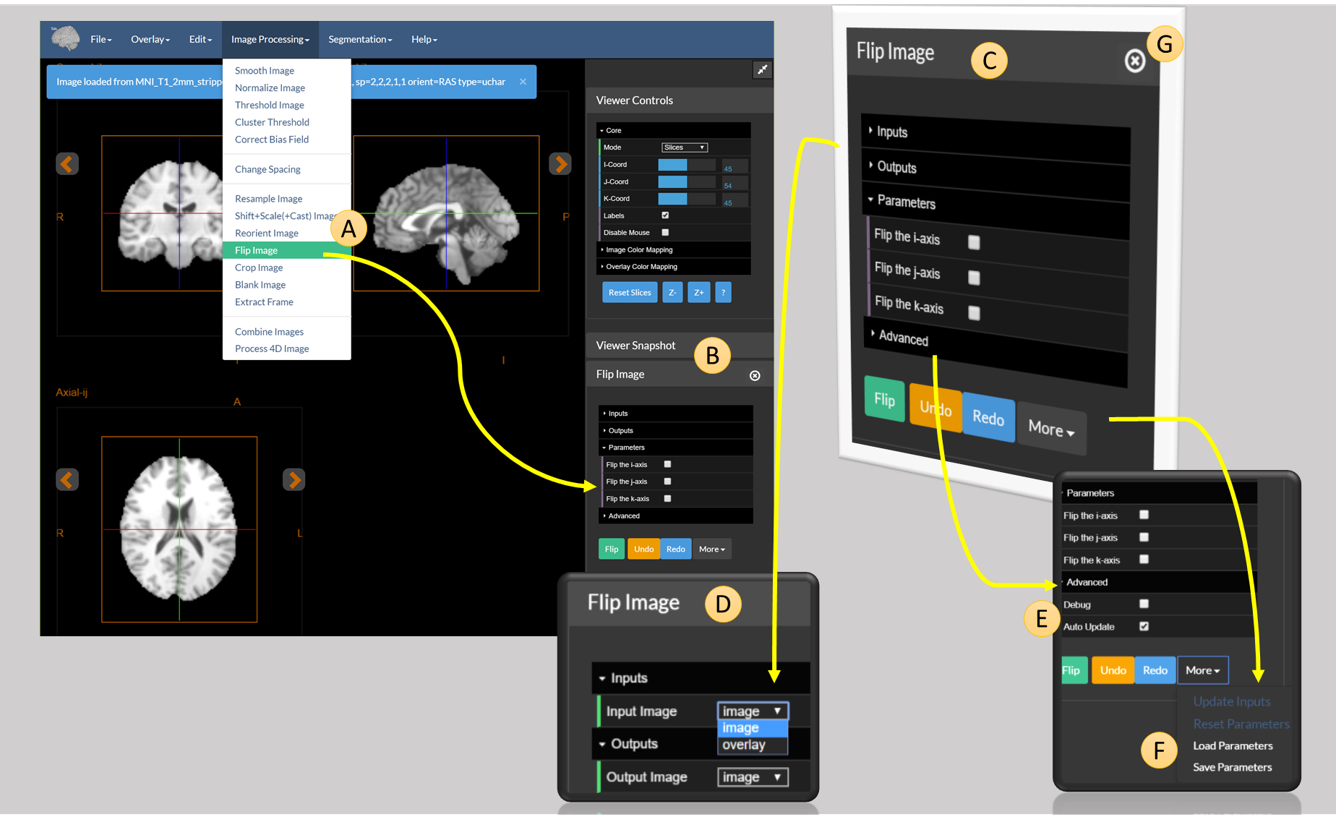 Figure 1: The full diagram explaining how to use a module in the viewer. (A) The Flip Image menu item (B) the Flip Image module in the sidebar (C) a cutaway of the Flip Image module (D) the inputs and outputs tab of Flip Image (E) advanced options for Flip Image (F) extra options for the module (G) the close button for the module in the sidebar.
Figure 1: The full diagram explaining how to use a module in the viewer. (A) The Flip Image menu item (B) the Flip Image module in the sidebar (C) a cutaway of the Flip Image module (D) the inputs and outputs tab of Flip Image (E) advanced options for Flip Image (F) extra options for the module (G) the close button for the module in the sidebar.
The tool is accessed using the Flip Image option under the Image Processing Menu (A). This opens the tool and places it in the viewer sidebar (B).
An expanded version of B is shown in C. The tool control consists of the following options:
- The
Inputstab (D). Allows the user to select which image to use as input. One of eitherimageoroverlay. - The
Outputstab. Similar to theInputstab, this determines whether to send the result of the module toimageoroverlay. - The
Parameterstab. Allows the user to set the parameters of the operation — in this case, the parameters allow the user to select which combination of the three axis (i,j, ork) to flip. - The
Advancedtab (E). Allows the user to set advanced parameters. These shouldn’t be needed in most cases, but are available for users with complex use cases. - The bar at the bottom contains three buttons:
- Flip — Runs the module.
- Undo and Redo — Undo or redo the last operation performed by the module. Note that this may not always work!
- The dropdown menu
Morewhich provides some more advanced operations (F).
Note: Outputs from a module will overwrite whatever is currently in their destination viewer. Be sure that you save your data as you go! Images can be moved from image to overlay and back, using the Advanced Transfer Tool, accessible under the Edit menu (see its section in the documentation).
The extra options under F merit some explanation. These are:
- Update Inputs — Used in certain advanced scenarios to force the module to reread its input from the viewer.
- Reset parameters — Resets the module to its default parameters.
-
Save Parameters — Saves the current parameters for the module to a JSON (
.param) file that takes the form:{ "module": "flipImage", "params": { "flipi": false, "flipj": true, "flipk": false, "debug": false } } - Load Parameters — Used to load back an existing parameter set and to set the values in the tool.
One advanced usage scenario is to configure parameter sets in the GUI (web-applications), especially for complex tasks, and to reuse these on the command line. The command line tools have a flag (—paramfile) that can be used to load all parameters from a such a parameter file and use them while running the module.
G highlights the close button. The sidebar can show only up to four tools (see the figure below). If an additional tool is opened, at this point, then the least recently opened module — in this case below Normalize Image — will be removed from the sidebar. The user may manually remove a control from the sidebar using the x button in its title bar.
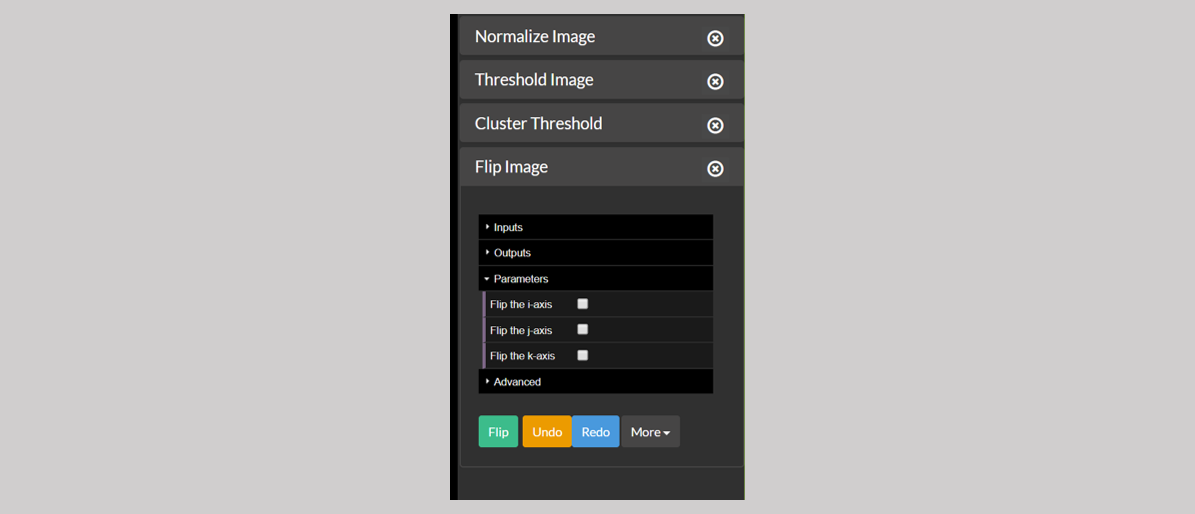 Figure 2: A sidebar full of modules.
Figure 2: A sidebar full of modules.
Using a Tool — Dual Viewer Mode
In the Dual Viewer application, there is an additional set of options for setting the input and the output image. As shown in Figure 3, both the input and the output have an additional “Viewer” tab with which you can choose to flip the image in viewer 1 and send the output to viewer 2 or any one the other possible combinations. The rest of the functionality is identical.
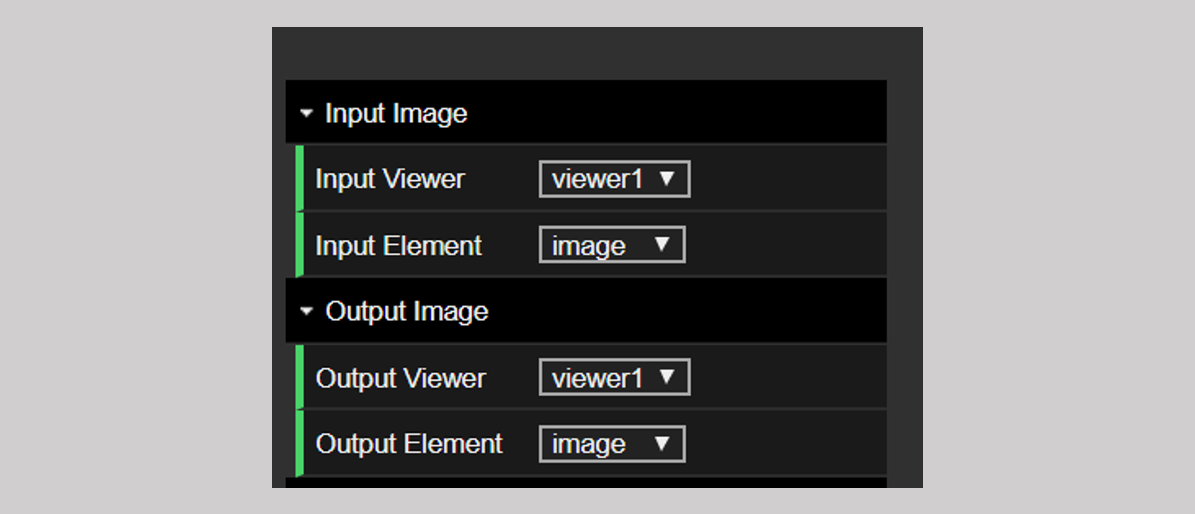 Figure 3: The input and output options in the dual viewer
Figure 3: The input and output options in the dual viewer
[BioImage Suite Web Manual Table Of Contents] [BioImage Suite Web Main Page]
This page is part of BioImage Suite Web. We gratefully acknowledge support from the NIH Brain Initiative under grant R24 MH114805 (Papademetris X. and Scheinost D. PIs, Dept. of Radiology and Biomedical Imaging, Yale School of Medicine.)
